Single-molecule Targeted Accessibility and Methylation Sequencing of Centromeres, Telomeres, and rDNAs in Arabidopsis
Abstract
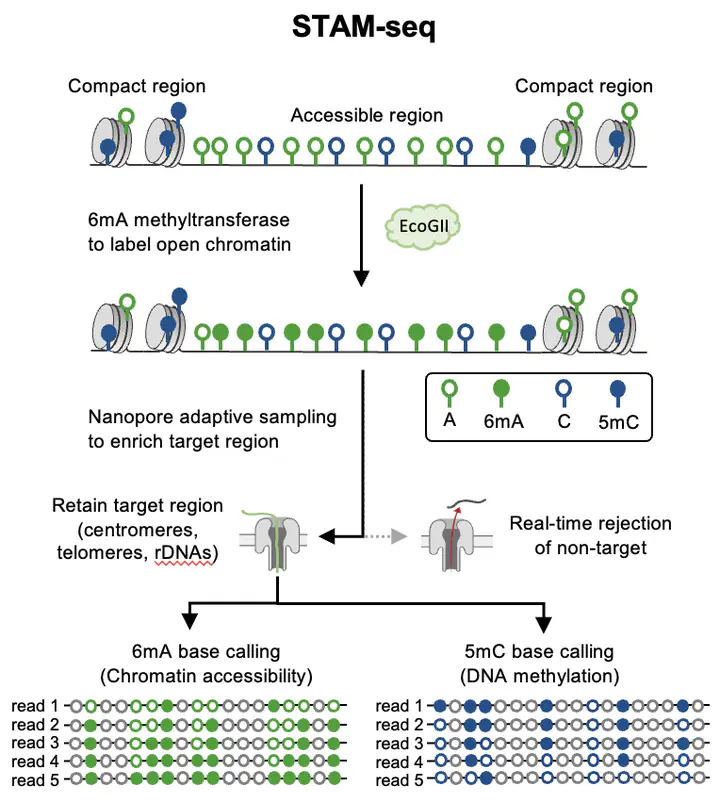
Next-generation sequencing (NGS) approaches have greatly expanded our understanding of epigenome, yet a precise characterization of highly repetitive regions (HRRs) remains challenging due to the short read-length of NGS that jeopardizes unique mapping at HRRs. Here, we developed a Nanopore-based long-read method STAM-seq (Single-molecule Targeted Accessibility and Methylation Sequencing) to specifically enrich and examine the epigenetic status of HRRs in Arabidopsis. Our single-molecule long-read analysis revealed that centromeres exhibit either accessible or inaccessible bipolar status; DNA methylation level of each individual rDNA unit is strongly anti-correlated with its accessibility in different variant groups; and telomere length is associated with subtelomeric DNA methylation. STAM-seq can be applied to study the accessibility and methylation of repetitive or high-copy sequences in a wide range of plant species.